GNN学习笔记
GNN从入门到精通课程笔记
3.1 GCN (ICLR ‘17)
- Semi-supervised Classification with Graph Convolutional Network (ICLR ‘17)
Abstract
- We present a scalable approach for semi-supervised learning on graph-structured data that is based on an efficient variant of convolutional neural networks which operate directly on graphs.
- We motivate the choice of our convolutional architecture via a localized first-order approximation of spectral graph convolutions.
- Our model scales linearly in the number of graph edges and learns hidden layer representations that encode both local graph structure and features of nodes.
Introduction
- Graph-based semi-supervised learning: classifying nodes in a graph network, where labels are only available for a small subset of nodes.
- f(X,A)
- X: a matrix of node feature vectors $X_i$
- A: an adjacency matrix
- Contribution:
- a simple and well-behaved layer-wise propagation rule for neural network models which operate directly on graphs
- how this form of a graph-based neural network model can be used for fast and scalable semi-supervised classification of nodes in a graph
Fast Approximate convolutions on graphs
- Basic Appoach
- Average neighbor messages and apply a neural network
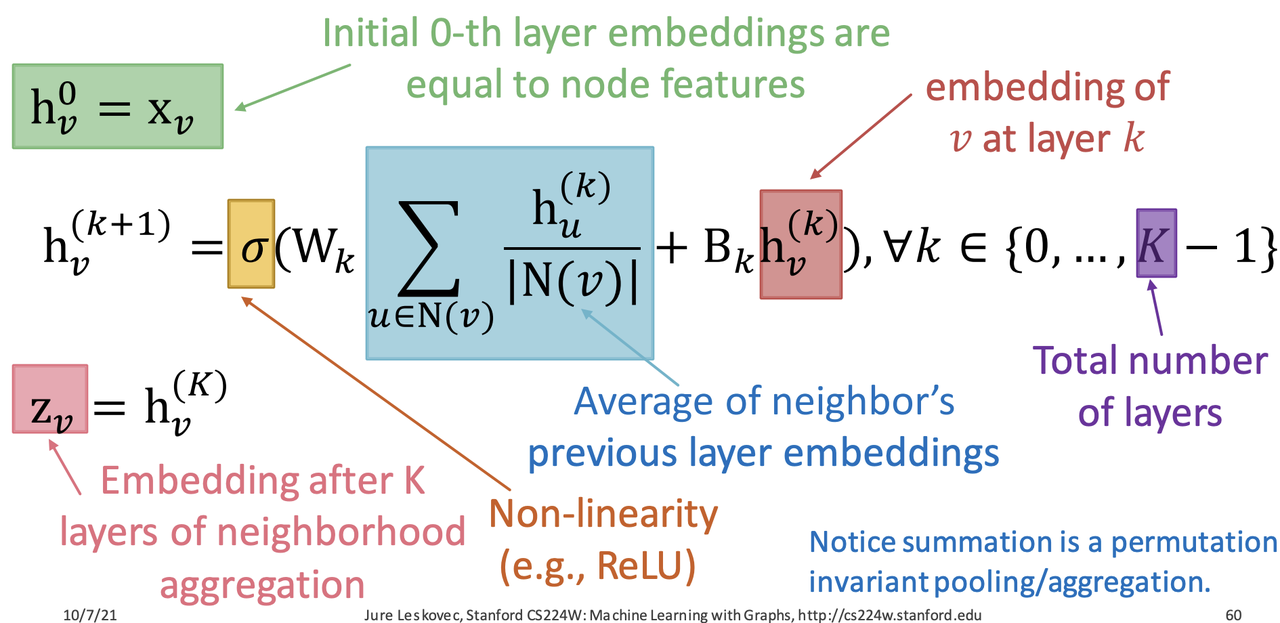
- Matrix Formulation: $H^{k+1} = \sigma(\tilde{A}H^kW_k^T+H^kB_k^T)$
- $\tilde{A} = D^{-1}A$: row normalized matrix
- $\tilde{A}H^kW_k^T$ : neighborhood aggregation
- $H^kB_k^T$: self transformation
- Average neighbor messages and apply a neural network
- Improvement
- The normalized adjacency matrix: $\tilde{A} = D^{-1/2}(A)D^{-1/2}$
- $\tilde{A} = A + A_N$, $\tilde{D}_{ii} = \Sigma{\tilde{A}_{ij}}$

- H: features
Semi-supervised Node Classification
- adjacency matrix: $\hat{A} = \tilde{D}^{-\frac{1}{2}}\tilde{A}\tilde{D}^{\frac{1}{2}}$
- Two-layer GCN for semi-supervised node classification
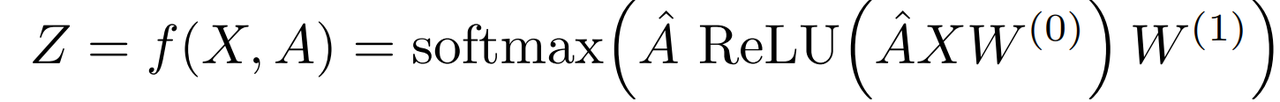
Experiments
- scales linearly in the number of graph edges
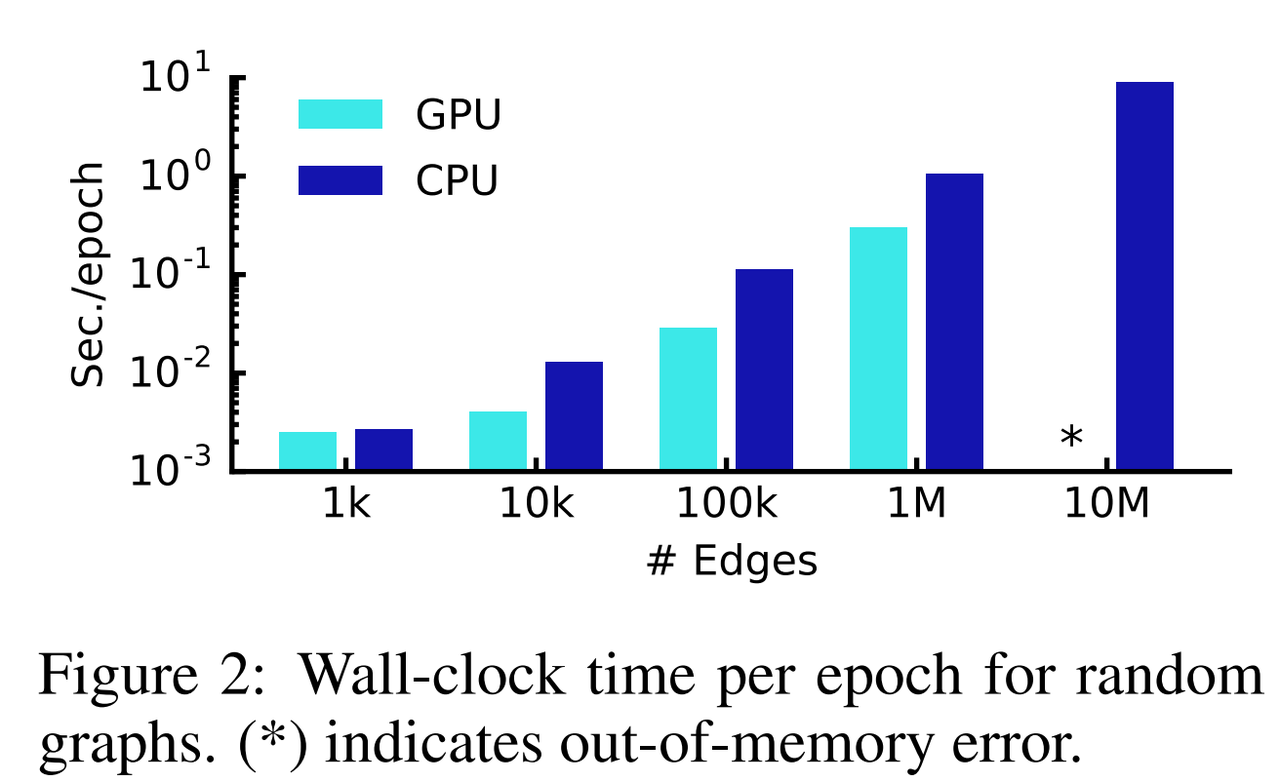
Implementation
- [1] Kipf, Thomas N., and Max Welling. “Semi-supervised classification with graph convolutional networks.” arXiv preprint arXiv:1609.02907 (2016).
- [2] https://github.com/tkipf/pygcn
1 | # graph preprocess |